Non-covalent interactions benchmark
[1]:
import pymolpro
import pandas as pd
[2]:
backend = 'local' # If preferred, change this to one of the backends in your ~/.sjef/molpro/backends.xml that is ssh-accessible
project_name = 'Non-covalent interactions benchmark'
parallel = None # how many jobs to run at once
[3]:
methods = {"HF": "df-hf", "MP2": "df-mp2", "LMP2": "df-lmp2", }
bases = ['aug-cc-pVDZ', 'aug-cc-pVTZ', 'aug-cc-pVQZ']
[4]:
db = pymolpro.database.load("GMTKN55_S22").subset('small')
[5]:
results = {}
for method in methods:
results[method] = {}
for basis in bases:
results[method][basis] = pymolpro.database.run(db, method=methods[method], basis=basis, location=project_name,
backend=backend, parallel=parallel)
[6]:
for method in methods:
for result in pymolpro.database.basis_extrapolate(results[method].values(),results['HF'].values()):
results[method][result.basis]=result
for basis in results[method]:
if basis not in bases: bases.append(basis)
[7]:
pd.set_option('display.precision', 2)
method_errors = pymolpro.database.analyse([results[method]['aug-cc-pVQZ'] for method in methods], db, 'kJ/mol')[
'reaction statistics']
with open(project_name + '.method_errors.tex', 'w') as tf:
tf.write('\\ifx\\toprule\\undefined\\def\\toprule{\\hline\\hline}\n\\def\\midrule{\\hline}\n\\def\\bottomrule{\\hline\\hline}\\fi') # or \usepackage{booktabs}
tf.write(method_errors.style.format(precision=2).to_latex(hrules=True,multicol_align='c',caption='Method errors'))
method_errors
[7]:
| DF-HF/aug-cc-pVQZ | DF-MP2/aug-cc-pVQZ | DF-LMP2/aug-cc-pVQZ | |
|---|---|---|---|
| MAD | 6.08 | 0.22 | 0.54 |
| MAXD | 7.52 | 0.42 | 0.79 |
| RMSD | 6.30 | 0.28 | 0.58 |
| MSD | -6.08 | 0.22 | -0.54 |
| STDEVD | 2.03 | 0.21 | 0.26 |
[8]:
for key, table in pymolpro.database.analyse([results[method]['aug-cc-pVQZ'] for method in methods], db).items():
print(key)
print(table)
results
[{'method': ['hf', 'df-hf'], 'basis': 'aug-cc-pVQZ', 'unit': 'Hartree', 'molecule energies': {'02': -152.137348921515, '08': -80.4318272687273, '01': -112.450005978623, '01a': -56.2239391353761, '02a': -76.0660246769283, '08a': -40.2162104675589}}, {'method': ['hf', 'df-mp2'], 'basis': 'aug-cc-pVQZ', 'unit': 'Hartree', 'molecule energies': {'02': -152.711896534593, '08': -80.8555325153533, '01': -112.960568865909, '01a': -56.4777418852842, '02a': -76.3518936483437, '08a': -40.4273483053424}}, {'method': ['hf', 'df-lmp2'], 'basis': 'aug-cc-pVQZ', 'unit': 'Hartree', 'molecule energies': {'02': -152.709533151809, '08': -80.8536174728202, '01': -112.958067503889, '01a': -56.4766434227825, '02a': -76.3509422922261, '08a': -40.4264399856976}}]
reaction energies
DF-HF/aug-cc-pVQZ DF-MP2/aug-cc-pVQZ DF-LMP2/aug-cc-pVQZ
2 5.30e-03 8.11e-03 7.65e-03
1 2.13e-03 5.09e-03 4.78e-03
8 -5.94e-04 8.36e-04 7.38e-04
reaction energy deviations
DF-HF/aug-cc-pVQZ DF-MP2/aug-cc-pVQZ DF-LMP2/aug-cc-pVQZ
2 -2.65e-03 1.59e-04 -3.02e-04
1 -2.87e-03 9.23e-05 -2.12e-04
8 -1.43e-03 -3.92e-06 -1.02e-04
reaction statistics
DF-HF/aug-cc-pVQZ DF-MP2/aug-cc-pVQZ DF-LMP2/aug-cc-pVQZ
MAD 2.32e-03 8.50e-05 2.05e-04
MAXD 2.87e-03 1.59e-04 3.02e-04
RMSD 2.40e-03 1.06e-04 2.21e-04
MSD -2.32e-03 8.24e-05 -2.05e-04
STDEVD 7.72e-04 8.18e-05 1.00e-04
molecule energies
DF-HF/aug-cc-pVQZ DF-MP2/aug-cc-pVQZ DF-LMP2/aug-cc-pVQZ
02 -152.14 -152.71 -152.71
08 -80.43 -80.86 -80.85
01 -112.45 -112.96 -112.96
01a -56.22 -56.48 -56.48
02a -76.07 -76.35 -76.35
08a -40.22 -40.43 -40.43
molecule violin plot
None
reaction violin plot
Figure(600x600)
[9]:
pd.set_option('display.precision', 2)
method_errors = pymolpro.database.analyse([results[method]['aug-cc-pVQZ'] for method in methods], db, 'kJ/mol')[
'reaction energy deviations']
method_errors
[9]:
| DF-HF/aug-cc-pVQZ | DF-MP2/aug-cc-pVQZ | DF-LMP2/aug-cc-pVQZ | |
|---|---|---|---|
| 2 | -6.96 | 0.42 | -0.79 |
| 1 | -7.52 | 0.24 | -0.56 |
| 8 | -3.76 | -0.01 | -0.27 |
[10]:
pd.set_option('display.precision', 2)
basis_errors = pymolpro.database.analyse([results['LMP2'][basis] for basis in bases], db, 'kJ/mol')[
'reaction statistics']
with open(project_name + '.basis_errors.tex', 'w') as tf:
tf.write('\\ifx\\toprule\\undefined\\def\\toprule{\\hline\\hline}\n\\def\\midrule{\\hline}\n\\def\\bottomrule{\\hline\\hline}\\fi') # or \usepackage{booktabs}
tf.write(basis_errors.style.format(precision=2).to_latex(hrules=True,multicol_align='c',caption='Basis errors'))
basis_errors
[10]:
| DF-LMP2/aug-cc-pVDZ | DF-LMP2/aug-cc-pVTZ | DF-LMP2/aug-cc-pVQZ | DF-LMP2/aug-cc-pV[23]Z | DF-LMP2/aug-cc-pV[34]Z | |
|---|---|---|---|---|---|
| MAD | 1.43 | 4.05 | 0.54 | 4.86 | 2.24 |
| MAXD | 2.23 | 10.63 | 0.79 | 13.76 | 6.46 |
| RMSD | 1.64 | 6.18 | 0.58 | 7.95 | 3.73 |
| MSD | -1.21 | -4.05 | -0.54 | -4.86 | 2.07 |
| STDEVD | 1.36 | 5.71 | 0.26 | 7.71 | 3.81 |
[11]:
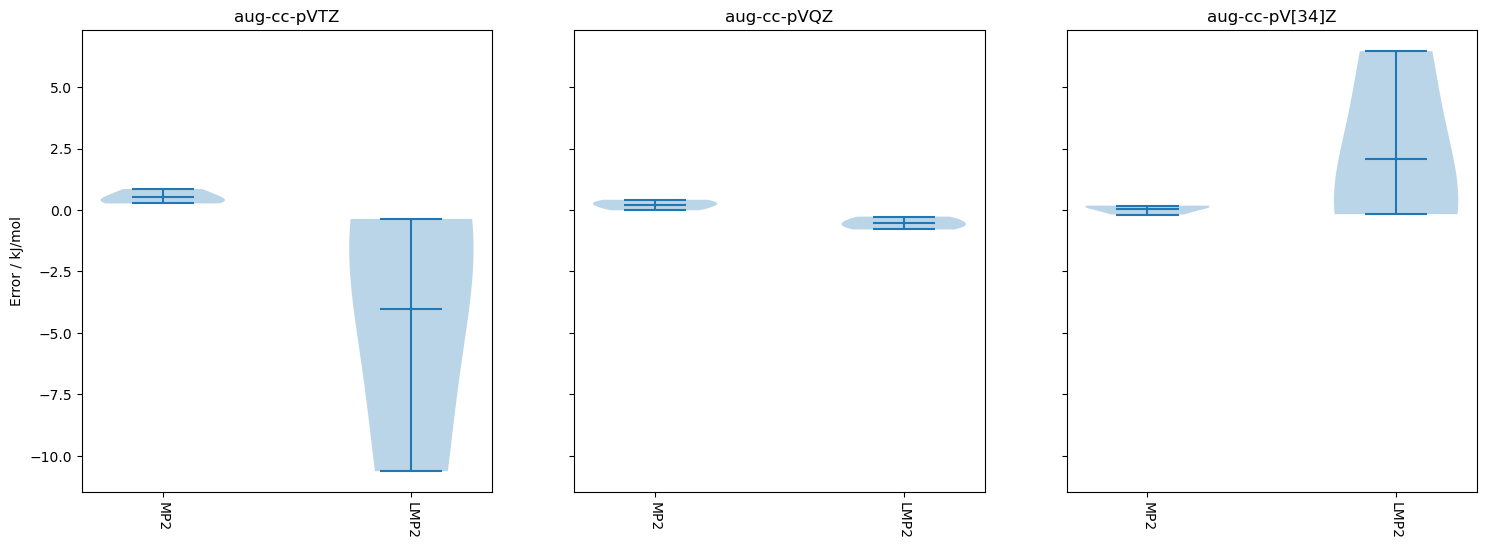
import matplotlib.pyplot as plt
methods_pruned = [method for method in methods if method != 'HF']
bases_pruned = ['aug-cc-pVTZ', 'aug-cc-pVQZ', 'aug-cc-pV[34]Z']
fig, panes = plt.subplots(nrows=1, ncols=len(bases_pruned), sharey=True, figsize=(18, 6))
for pane in range(len(bases_pruned)):
data = []
for method in methods_pruned:
data.append(
pymolpro.database.analyse(results[method][bases_pruned[pane]],
db,'kJ/mol')['reaction energy deviations'].to_numpy()[:, 0]
)
panes[pane].violinplot(data, showmeans=True, showextrema=True, vert=True, bw_method='silverman')
panes[pane].set_xticks(range(1, len(methods_pruned) + 1), labels=methods_pruned, rotation=-90)
panes[pane].set_title(bases_pruned[pane])
panes[0].set_ylabel('Error / kJ/mol')
plt.savefig(project_name + ".violin.pdf")
[12]:
basis_errors.to_latex(float_format='%.2f')
[12]:
'\\begin{tabular}{lrrrrr}\n\\toprule\n & DF-LMP2/aug-cc-pVDZ & DF-LMP2/aug-cc-pVTZ & DF-LMP2/aug-cc-pVQZ & DF-LMP2/aug-cc-pV[23]Z & DF-LMP2/aug-cc-pV[34]Z \\\\\n\\midrule\nMAD & 1.43 & 4.05 & 0.54 & 4.86 & 2.24 \\\\\nMAXD & 2.23 & 10.63 & 0.79 & 13.76 & 6.46 \\\\\nRMSD & 1.64 & 6.18 & 0.58 & 7.95 & 3.73 \\\\\nMSD & -1.21 & -4.05 & -0.54 & -4.86 & 2.07 \\\\\nSTDEVD & 1.36 & 5.71 & 0.26 & 7.71 & 3.81 \\\\\n\\bottomrule\n\\end{tabular}\n'
[13]:
print(basis_errors.style.format(precision=3).to_latex(hrules=True,multicol_align='c'))
\begin{tabular}{lrrrrr}
\toprule
& DF-LMP2/aug-cc-pVDZ & DF-LMP2/aug-cc-pVTZ & DF-LMP2/aug-cc-pVQZ & DF-LMP2/aug-cc-pV[23]Z & DF-LMP2/aug-cc-pV[34]Z \\
\midrule
MAD & 1.435 & 4.047 & 0.539 & 4.857 & 2.239 \\
MAXD & 2.228 & 10.627 & 0.793 & 13.761 & 6.464 \\
RMSD & 1.642 & 6.175 & 0.580 & 7.952 & 3.734 \\
MSD & -1.207 & -4.047 & -0.539 & -4.857 & 2.071 \\
STDEVD & 1.364 & 5.712 & 0.262 & 7.712 & 3.805 \\
\bottomrule
\end{tabular}